Transmission Electron Microscopy (TEM)
TEM utilizes a beam of highly energized electrons that transmit through a thin sample to form an image that reveals its fine details, with (sub)nanometer spatial resolution. This technique finds wide applications in the analysis of various biological samples, including (bio)nanomaterials, cells, and tissue sections. At Radboudumc EMC, we are dedicated to providing state-of-the-art TEM imaging services. Our facility is equipped with both standard and advanced modalities, ensuring that we offer cutting-edge capabilities to meet a wide range of research needs. We invite you to explore the possibilities of TEM imaging available at our center.
Our TEM instrument
Our center is equipped with a Thermo Fisher TALOS F200C-G2 microscope specifically designed for beyond-state-of-art TEM imaging.
- a phase plate system for enhanced contrast in TEM at resolutions > 1 nm;
a Falcon 4i Direct electron detector camera;
- segmented STEM detector for differential phase contrast imaging in STEM;
- an energy-dispersive X-ray spectroscopy (EDS) system for elemental analysis;
- a LED light source mounted in the column, to provide light pulses for initiating processes in the microscope
Cryogenic TEM
Cryo-TEM of plunge-frozen samples
For
(bio)nanostructures, like extracellular vesicles or protein (nano)particles,
sample preparation for cryo-TEM is straightforward and can be done with
our Vitrobot automated plunge freezing system.
In plunge freezing, a small droplet of the liquid dispersion containing the
structures to be observed is applied to a TEM grid. The grid with the sample is
then rapidly plunged into a cryogen (liquid ethane), at temperatures below
-180°C. The rapid freezing process ensures the sample is vitrified into an
amorphous ice state, preserving the native structural and
molecular details of the studied materials. We
used cryoTEM to study a range of
(bio)nanomaterials, such as extracellular vesicles, collagen fibrils, drug
delivery vehicles, particulate biomaterials and mineral particles.
Moreover, we perform cryoTEM imaging
in both 2D and 3D (cryo-electron
tomography,
cryoET).
Cryo-TEM to observe cells and tissues in a near-native state
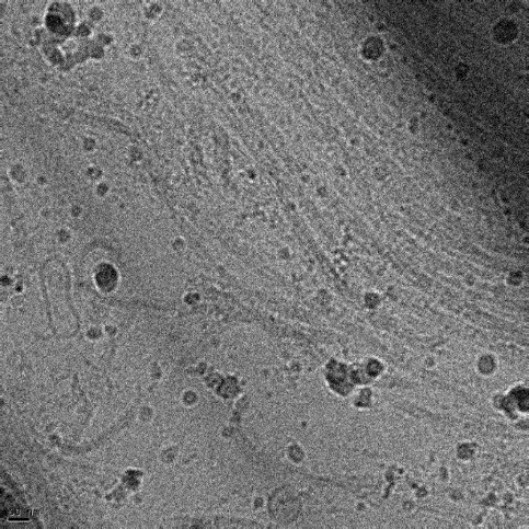
Cryo-TEM
is not only suitable for observing (bio)nanostructures in aqueous environments
but also for revealing ultrastructural details of cells and tissues in a
near-native state. However, individual cells or tissues are too thick for the
electron transparency required in TEM imaging. To address this challenge, we
have developed a specialized workflow. When high-resolution cryo-TEM
imaging is needed for thick samples, we first employ high-pressure freezing to
safely vitrify the sample rapidly. Then, we
use a focused ion beam (FIB) to create
a thin lamella from
the vitrified sample we can
lift out for cryo-TEM imaging. This approach allows for 2D
or 3D (cryoET) visualization of cellular
details, within their natural context and in their native
hydrated state. With our workflow for correlative light and electron microscopy,
we can precisely identify and localize
fluorescently-labeled targets for high-resolution
cryo-TEM imaging.
Liquid Phase EM
We are the national facility for liquid phase EM of biological materials
Liquid Phase (LP) EM is an imaging method that enables the direct visualization of samples immersed in their liquid environments with nanometer spatial resolution and (sub-)second temporal resolution. LP-EM has been used to observe dynamic processes at the nanoscale, such as the self-assembly of polymeric vesicles (see video). The capability to capture events in real-time makes LP-EM a promising approach that could revolutionize how we study biological processes.
So far, the bottleneck in the application of LP-EM for imaging biological materials has been the damaging effect of repeated electron beam radiation, which may alter the structure of the beam sensitive mateials and alter (bio)chemical processes within the sample.
To minimize the risk on damaging the sample during imaging, we have established a state-of-the-art facility dedicated to LP-EM imaging of low-constrast biological materials. This facility incorporates cutting-edge experimental configurations for different samples, that optimize acquisition rates through the use of fast and sensitive detectors, and enhance sample contrast.
Real-time imaging of the self-assembly of polymeric nanovesicles
Different experimental configurations
We have set-ups to perform LP-EM experiments for different types of samples and experimental configurations.
Liquid Flow cells with temperature control. We have a Denssolutions Stream liquid cell holder, in which the liquid sample enclosed within two microchips that allows the application of flow with high control over flow conditions and temperature.
Graphene liquid cells. We use an automated Vitrotem Naiad system for the encapsulation of liquid samples of biological materials in between two sheets of graphene. The ultra-thin graphene layers function as electron-transparent windows, enabling high-resolution imaging while mitigating electron beam damage. We are also currently working on developing methods to cryo-arrest processes into the liquid pockets and reinitiating them inside the TEM for dynamic imaging.
In situ fluorescence. Our TEM instrument is equipped with a world-wide unique light source mounted inside the column, to provide light pulses for initiating processes inside the microscope, in particular within graphene liquid cells..
National Facility: BIOMATEM
The establishment of a unique national facility for LP-EM of biological materials is supported by the NWO Investment Grant Large (NWO Groot) project BIOMATEM and a financial contribution from Radboudumc. BIOMATEM brings together a consortium of biologists, (bio)chemists, physicists, and materials scientists all working towards in situ nanoscale imaging to understand biological and biomedical processes. The team covers all aspects, from developing sample preparation methodologies, to developing low-dose imaging strategies, and the application of LP-EM for studying biological processes, from fundamental as well as clinically relevant perspectives. In the upcoming years, different projects involving LP-EM of biological materials will be developed in our facility by different groups from the Netherlands, addressing amongst others the following topics:
- Collagen mineralization process in health and disease by Prof. Nico Sommerdijk's group (Radboudumc).
- Visualization of vesicular membrane fusion by Prof. Alexander Kros's group (Leiden University)
- Targeted drug delivery with self-propelling nanoparticles by Prof. Daniela Wilson's group (Radboud University)
- Vesicular trafficking and fusion in human neurons by Prof. Matthijs Verhage's group (Vrije Universiteit Amsterdam)
- Intracellular drug delivery from protein nanoparticles by Prof. Sander Leeuwenburgh's group (Radboudumc)
Room Temperature TEM
TEM imaging of resin-embedded cells
TEM is routinely used to reveal ultrastructural details of cells and tissues. This involves a sample preparation step to obtain thin, electron transparent, sections (<100 nm) of the cells and tissues. A typical workflow involves three steps: fixing and dehydrating the specimen, embedding it in resin, and cutting ultra-thin sections that usuallu are less than 100 nm thick. The key to a successful experiment is the sample preparation that must assure the preservation of the ultrastructural features of interest. At Radboudumc EMC, we offer different approaches for preparing cells and tissues for standard TEM imaging that are tailored to maximize the results.
Chemical Fixation and resin-embedding
The most basic approach is to use a chemical agent (e.g. glutaraldehyde) to fix the sample and preserve its structure. Next, the samples is stained with heavy metal, dehydrated, and embedded in resin. Using our microtome (Leica Artros 3D ultramicrotome) the resin block is carefully sliced into thin (<100 nm) sections which are then placed on a TEM grid for imaging. In this way we can visualize cell membranes and structural details of cell organelles, as well as the extracellular matrix.
High-pressure freezing and freeze substitution
However, the chemical fixation of cells can introduce artifacts that affect the fidelity of the samples ultrastructure. Near-native preservation of the native cellular ultrastructure is achieved by cryogenic fixation of sample up to 200 micrometer in thickness through high-pressure freezing (HPF). HPF freezes the sample in 2 microseconds by exposing it to liquid nitrogen at 2000 atmosphere, stopping all processes and trapping all structures within a vitreous ice state. Then, the samples can be embedded in resin using freeze substitution replacing water within a frozen sample with a cryoprotective solution. By slowly warming the sample while maintaining sub-zero temperatures, freeze substitution allows for the removal of ice crystals and the infiltration of the cryoprotective solution. We have successfully employed this approach to observe Covid virus particles within cells of kidney organoids.
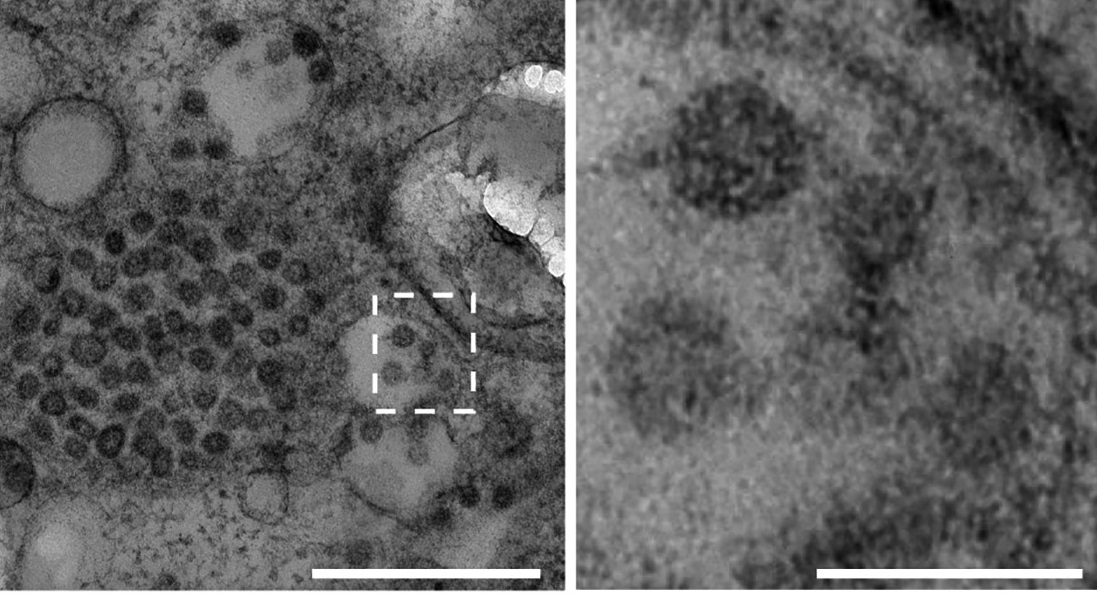
TEM ultrastructure analysis of viral particles (60–70 nm) in the corresponding SARS-CoV-2 nucleocapsid protein-positive cell within the kidney organoid. Scale bars, (left) 500 nm; (right) scale bars, 100 nm.


